- About
- Mission Statement
Education. Evidence. Regrowth.
- Education.
Prioritize knowledge. Make better choices.
- Evidence.
Sort good studies from the bad.
- Regrowth.
Get bigger hair gains.
Team MembersPhD's, resarchers, & consumer advocates.
- Rob English
Founder, researcher, & consumer advocate
- Research Team
Our team of PhD’s, researchers, & more
Editorial PolicyDiscover how we conduct our research.
ContactHave questions? Contact us.
Before-Afters- Transformation Photos
Our library of before-after photos.
- — Jenna, 31, U.S.A.
I have attached my before and afters of my progress since joining this group...
- — Tom, 30, U.K.
I’m convinced I’ve recovered to probably the hairline I had 3 years ago. Super stoked…
- — Rabih, 30’s, U.S.A.
My friends actually told me, “Your hairline improved. Your hair looks thicker...
- — RDB, 35, New York, U.S.A.
I also feel my hair has a different texture to it now…
- — Aayush, 20’s, Boston, MA
Firstly thank you for your work in this field. I am immensely grateful that...
- — Ben M., U.S.A
I just wanted to thank you for all your research, for introducing me to this method...
- — Raul, 50, Spain
To be honest I am having fun with all this and I still don’t know how much...
- — Lisa, 52, U.S.
I see a massive amount of regrowth that is all less than about 8 cm long...
Client Testimonials150+ member experiences.
 Scroll DownPopular Treatments
Scroll DownPopular Treatments- Treatments
Popular treatments. But do they work?
- Finasteride
- Oral
- Topical
- Dutasteride
- Oral
- Topical
- Mesotherapy
- Minoxidil
- Oral
- Topical
- Ketoconazole
- Shampoo
- Topical
- Low-Level Laser Therapy
- Therapy
- Microneedling
- Therapy
- Platelet-Rich Plasma Therapy (PRP)
- Therapy
- Scalp Massages
- Therapy
More
IngredientsTop-selling ingredients, quantified.
- Saw Palmetto
- Redensyl
- Melatonin
- Caffeine
- Biotin
- Rosemary Oil
- Lilac Stem Cells
- Hydrolyzed Wheat Protein
- Sodium Lauryl Sulfate
More
ProductsThe truth about hair loss "best sellers".
- Minoxidil Tablets
Xyon Health
- Finasteride
Strut Health
- Hair Growth Supplements
Happy Head
- REVITA Tablets for Hair Growth Support
DS Laboratories
- FoliGROWTH Ultimate Hair Neutraceutical
Advanced Trichology
- Enhance Hair Density Serum
Fully Vital
- Topical Finasteride and Minoxidil
Xyon Health
- HairOmega Foaming Hair Growth Serum
DrFormulas
- Bio-Cleansing Shampoo
Revivogen MD
more
Key MetricsStandardized rubrics to evaluate all treatments.
- Evidence Quality
Is this treatment well studied?
- Regrowth Potential
How much regrowth can you expect?
- Long-Term Viability
Is this treatment safe & sustainable?
Free Research- Free Resources
Apps, tools, guides, freebies, & more.
- Topical Finasteride Calculator
- Interactive Guide: What Causes Hair Loss?
- Free Guide: Standardized Scalp Massages
- 7-Day Hair Loss Email Course
- Ingredients Database
- Interactive Guide: Hair Loss Disorders
- Treatment Guides
- Product Lab Tests: Purity & Potency
- Evidence Quality Masterclass
More
Articles100+ free articles.
-
Cannabidiol (CBD) Increases Hair Counts By 246%? Not So Fast.
-
Creatine: Does It Worsen Hair Loss? It Depends On The Hair Loss Type.
-
Can Progesterone Improve Hair Regrowth?
-
CRABP2: Can This Gene Predict Regrowth From Retinoids?
-
BTD: Can This Gene Predict Regrowth From Biotin?
-
COL1A1: Can This Gene Predict Regrowth From Collagen Support?
-
2dDR For Hair Loss: What Do We Know So Far About This Sugar?
-
CYP19A1: Can This Gene Predict Regrowth From Hormone Therapy?
PublicationsOur team’s peer-reviewed studies.
- Microneedling and Its Use in Hair Loss Disorders: A Systematic Review
- Use of Botulinum Toxin for Androgenic Alopecia: A Systematic Review
- Conflicting Reports Regarding the Histopathological Features of Androgenic Alopecia
- Self-Assessments of Standardized Scalp Massages for Androgenic Alopecia: Survey Results
- A Hypothetical Pathogenesis Model For Androgenic Alopecia:Clarifying The Dihydrotestosterone Paradox And Rate-Limiting Recovery Factors
Menu- AboutAbout
- Mission Statement
Education. Evidence. Regrowth.
- Team Members
PhD's, resarchers, & consumer advocates.
- Editorial Policy
Discover how we conduct our research.
- Contact
Have questions? Contact us.
- Before-Afters
ArticlesSRD5A1 & 2: Can These Genes Predict Regrowth From 5-Alpha-Reductase Inhibitors?
First Published Aug 6 2024Last Updated Oct 29 2024Pharmaceutical Researched & Written By:Ben Fletcher, PhD
Researched & Written By:Ben Fletcher, PhD Reviewed By:Rob English, Medical Editor
Reviewed By:Rob English, Medical Editor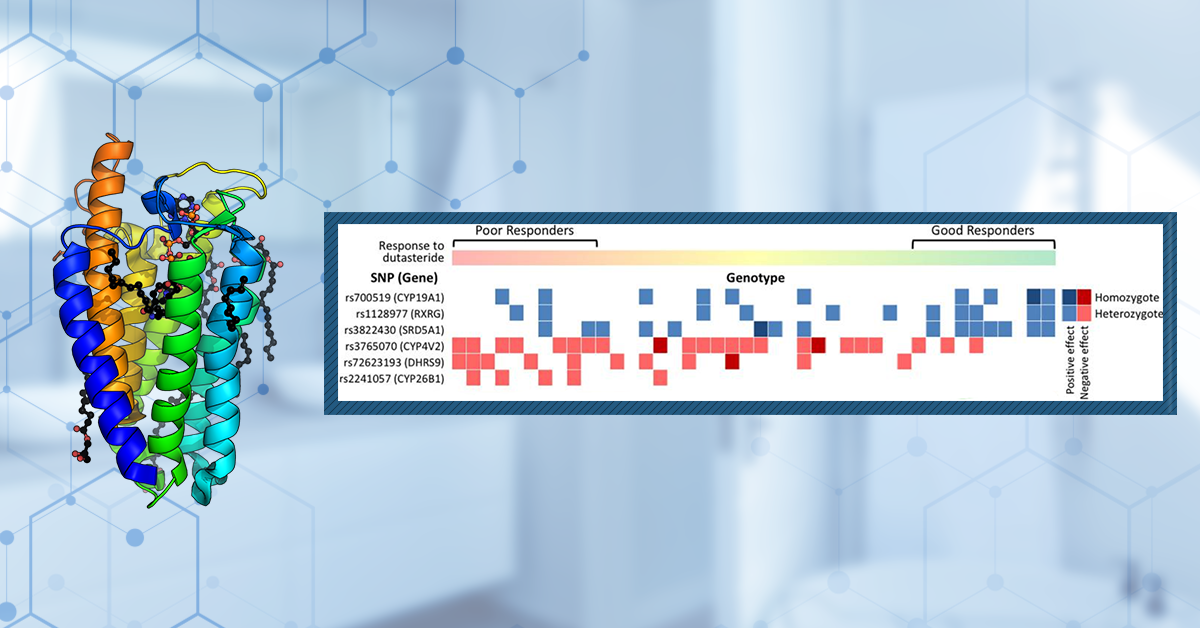
Want help with your hair regrowth journey?
Get personalized support, product recommendations, video calls, and more from our researchers, trichologists, and PhD's dedicated to getting you the best possible outcomes.
Learn MoreArticle Summary
Can your genes predict the effectiveness of 5α-reductase inhibitors for hair loss? The SRD5A1 and SRD5A2 genes, encoding type I and type II 5α-reductase respectively, are central to DHT production, a key factor in the pathogenesis of androgenic alopecia. Genetic testing companies suggest certain SR5DA1 and 2 variants might predict how well someone responds to treatments like finasteride and dutasteride. While early studies indicate potential links, these studies often have small sample sizes and varied methodologies. In this article, we explore the role of SRD5A1 and SRD5A2 in hair loss treatment and evaluate whether genetic variations can truly determine treatment success.
Full Article
SRD5A1 and SRD5A2 are both part of the SRD5A gene family, encoding type I and type II 5α-reductase, respectively. With regard to hair loss, 5α-reductase is a type of enzyme that catalyzes the conversion of testosterone into dihydrotestosterone (DHT). DHT is a driving factor in androgenic alopecia (AGA) and, as a result, 5α-reductase inhibitors are a primary treatment for AGA. A collection of studies have investigated genetic variation in SRD5A1 and SRD5A2, with the results suggesting that some variants may cause differential responses to treatment with 5α-reductase inhibitors. This article will explore how relevant SRD5A1 and SRD5A2 are to hair treatment effectiveness and how to interpret your genetic results to make the correct treatment choice.
What Are the SR5DA Genes?
The Steroid 5α-Reductase (SRD5A) gene family is thought to be composed of up to five genes, the most prominent being SRD5A1, SRD5A2, and SRD5A3. Despite the name, only SRD5A1 and SRD5A2 exhibit strong 5α-reductase activity, meaning that they are highly efficient at converting testosterone to DHT.[1]Scaglione, A., Montemiglio, L.C., Parisi, G., Asteriti, I.A., Bruni, R., Cerutti, G., Testi, C., Savino, C., Mancia, F., Lavia, P. and Vallone, B. (2017). Subcellular localization of the five members … Continue reading
Both SRD5A1 and SRD5A2 are expressed throughout the body, but it is their expression in the scalp that is important for hair growth outcomes. Type I 5α-reductase (produced by SRD5A1) is only strongly expressed in the sebaceous glands, the part of the hair follicle that produces sebum and keeps the skin moisturized. In contrast, type II 5α-reductase (produced by SRD5A2) is primarily expressed in the infundibulum (upper portion), inner root sheath, and outer root sheath of the hair follicle, indicating that type I and type II 5α-reductase have distinct localization within the skin and hair follicle.[2]Bayne, Flanagan, Einstein, Ayala, Chang, Azzolina, Whiting, Mumford and Thiboutot. (1999). Immunohistochemical localization of types 1 and 2 5α‐reductase in human scalp. British Journal of … Continue reading
Although they do have other roles, the primary function of these enzymes is to convert testosterone into dihydrotestosterone (DHT). One of four androgens (steroid hormones), DHT is known to be a driving factor of androgenic alopecia (AGA) and is implicated in the miniaturization of hair follicles.[3]Marchetti, P.M. and Barth, J.H. (2013). Clinical biochemistry of dihydrotestosterone. Annals of Clinical Biochemistry, 50(2), 95-107. Available at: https://doi.org/10.1258/acb.2012.012159
Due to the aforementioned role of DHT, 5α-reductase is a key target when it comes to the treatment of AGA. Thus, one of the main classes of AGA and hair loss treatments is 5α-reductase inhibitors. These inhibitors work by targeting the 5α-reductase enzymes, reducing their levels in order to disrupt the conversion of testosterone to DHT. Finasteride is the only FDA-approved 5α-reductase inhibitor for the treatment of AGA, which inhibits type II 5α-reductase. Dutasteride, a dual-inhibitor of type I and type II 5α-reductase, is only FDA-approved for the treatment of an enlarged prostate but is also prescribed off-label for AGA.[4]Gupta, A.K., Bamimore, M.A., Wang, T. and Talukder, M. (2024). The impact of monotherapies for male androgenetic alopecia: A network meta‐analysis study. Journal of Cosmetic Dermatology. Available … Continue reading
Other 5α-reductase inhibitors exist, including naturally occurring substances such as saw palmetto and some green tea catechins, but they have varying levels of evidence quality.
What is The Evidence For Targeting SR5DA for Hair Loss?
So, SRD5A1 and SRD5A2 are closely linked to 5α-reductase activity, DHT production, and, therefore, AGA. Owing to this, it is feasible that genetic variation in either SRD5A1 or SRD5A2 could influence the efficacy of 5α-reductase inhibitors in the treatment of hair loss.
In one study, the genotypes of 42 males with AGA were investigated. They had been using dutasteride for 6 months in a clinical trial, which had produced variable results. For the purposes of this study, patients were put into the poor responder (control) group or the good responder (cases) group, based on their increase in hair count during the original study. In their investigation, they identified a number of genetic variants that were statistically significantly associated with the strength of the response to the treatment (Figure 1).[5]Rhie, A., Son, H.Y., Kwak, S.J., Lee, S., Kim, D.Y., Lew, B.L., Sim, W.Y., Seo, J.S., Kwon, O., Kim, J.I. and Jo, S.J. (2019). Genetic variations associated with response to dutasteride in the … Continue reading
Three of these were single nucleotide polymorphisms (SNPs) within the SRD5A1 gene – rs3822430, rs8192186, and rs3736316. Each of these SNPs was positively associated with the response to dutasteride; in other words, patients with these genetic variants responded better to dutasteride treatment.

Figure 1: Genotypic landscape of 42 patients and the cumulative effect of each allele count and their positive or negative effect. Boxes represent SNPs that exhibited a positive (blue) or negative (red) effect on dutasteride response. Light-colored boxes represent heterozygous SNPs (a variation where an individual has two different versions of a specific DNA sequence at a particular location in the genome), dark-colored boxes represent homozygous SNPs (a variation where an individual has two identical versions of a specific DNA sequence at a particular location in the genome). Patient responses to dutasteride improve from left to right.[6]Rhie, A., Son, H.Y., Kwak, S.J., Lee, S., Kim, D.Y., Lew, B.L., Sim, W.Y., Seo, J.S., Kwon, O., Kim, J.I. and Jo, S.J. (2019). Genetic variations associated with response to dutasteride in the … Continue reading
Despite the positive associations, some of the patients who possessed these SNPs were still considered to be poor responders. This suggests that having a ‘positive’ SNP in SRD5A1 does not provide a definitive indication that you will respond well to dutasteride. Rather, the study’s results suggest that the combination of SNPs you possess and how they interact with each other is more important.
Interestingly, SRD5A2 was not associated with the response to dutasteride in the same study and no other studies have investigated the effects of SRD5A2 variants on the efficacy of dutasteride or finasteride treatment for hair loss, but that does not mean there are no effects.
Genetic variation in the SRD5A gene family appears to influence the efficacy of dutasteride treatment, so the question then turns to why this might happen.
In a study conducted on 57 males with type II diabetes, an association was found between the SRD5A1 HinfI SNP and the DHT/testosterone ratio, which is used as an indicator of 5α-reductase enzyme activity.[7]Ellis, J.A., Panagiotopoulos, S., Akdeniz, A., Jerums, G. and Harrap, S.B. (2005). Androgenic correlates of genetic variation in the gene encoding 5α-reductase type 1. Journal of Human Genetics, … Continue reading
HinfI is an enzyme that binds to a specific series of bases (called a restriction site) and cuts the DNA. Any changes to that sequence of bases will prevent HinfI from cutting the DNA, so it can be used to identify SNPs within known restriction sites. Individuals who lacked the SRD5A1 HinfI restriction site had a higher ratio of DHT/testosterone ratio than those with other genotypes, indicating increased activity of 5α-reductase.
In patients with increased levels of DHT, due to genetic variation, treatment with 5α-reductase inhibitors may look different. Higher concentrations of 5α-reductase inhibitors may be necessary, and it might take longer to see results, but the treatment may also have a more noticeable effect on hair loss as DHT levels can be reduced more than normal.

Figure 2: Measurements of testosterone (T), DHT, sex hormone binding globulin (SHBG), and insulin in participants with different genotypes (AA, AB, BB). A=absence of restriction site. Presence of the restriction site. Those lacking the restriction site (AA) had a higher DHT/T ratio meaning increased 5a-reductase activity.[8]Ellis, J.A., Panagiotopoulos, S., Akdeniz, A., Jerums, G. and Harrap, S.B. (2005). Androgenic correlates of genetic variation in the gene encoding 5α-reductase type 1. Journal of Human Genetics, … Continue reading
A different study was conducted on 104 patients with metastatic prostate cancer, investigating SNPs in SRD5A1 and SRD5A2. The authors discovered that patients with the GG genotype in SRD5A2 rs523349 exhibited increased 5α-reductase enzyme activity than those with the GC or CC genotype. #
The patients did not have their DHT levels measured, however, it is possible that increased 5α-reductase enzyme activity would lead to increased levels of DHT, which might make patients with the GG genotype a better candidate for treatment with a 5α-reductase inhibitor.[9]Shiota, M., Fujimoto, N., Yokomizo, A., Takeuchi, A., Itsumi, M., Inokuchi, J., Tatsugami, K., Uchiumi, T. and Naito, S. (2015). SRD5A gene polymorphism in Japanese men predicts prognosis of … Continue reading
These findings were supported by another study, which investigated several genetic variants of the SRD5A2 gene in healthy men. The authors found that, although the expression levels of the type II 5α-reductase enzyme were similar throughout, some of the variants produced type II 5α-reductase with very different properties to the normal (wild-type) enzyme.[10]Makridakis, N.M., di Salle, E. and Reichardt, J.K. (2000). Biochemical and pharmacogenetic dissection of human steroid 5α-reductase type II. Pharmacogenetics and Genomics, 10(5), 407-413. Available … Continue reading
Importantly, the activity of the enzyme (the speed at which it catalyzes reactions) was lower than normal in some variants and higher in others suggesting that DHT production may be lower in people with some SRD5A2 variants and higher in people with others. In contrast to those with higher levels, patients with lower DHT levels than normal may require a reduced dose of 5α-reductase inhibitor, and it is possible that treatment will be less efficient as DHT levels are already low. Or, other treatments that don’t primarily target DHT might be more appropriate.
To investigate this further, the authors produced lab-grown animal cells which contained each of the genetic variants. They then tested finasteride and dutasteride on all of the cells and measured 5α-reductase activity, to determine how efficiently the 5α-reductase inhibitors worked on each variant. They found that finasteride and dutasteride could be more, less, or equally effective as normal, depending on the variant(s).
Moreover, finasteride could be more effective than normal against a variant, while dutasteride was less effective than normal, and vice versa. This suggests that genetic variation could significantly affect the outcomes of treatment with finasteride or dutasteride.
Collectively, the results of these studies suggest that genetic variation in the SRD5A family of genes should be considered when designing treatment regimens for hair loss. However, the data is not without its limitations.
For the most part, studies conducted on SRD5A (including the ones here) have only included male participants. As males and females display differences in DHT expression, it would not be surprising if there were also differences in SRD5A gene expression and 5α-reductase inhibitor activity.
Furthermore, these studies were only conducted on a small number of participants. This reduces the power of a study, as you may not have an accurate representation of the wider population, and it can lead to results being misleading. Repeating the study on a much larger scale would produce more conclusive results. Also, some of the studies were not conducted on patients with AGA or hair loss-related conditions, so the findings are not directly relevant to hair loss treatment.
The final study, which investigated 5α-reductase inhibitor activity on each variant, was performed in vitro. In other words, it was performed on cells that had been genetically modified to express each variant. Furthermore, these cells were not even human cells found in the scalp or hair follicle, but rather animal cells.
Although models such as this can be very useful, the results they produce are not definitive, as the model (cells, in this case) is not a true representation of human anatomy.
Despite these limitations, there is sufficient evidence to suggest that genetic variation in SRD5A1 or SRD5A2 may influence your response to treatment with a 5α-reductase inhibitor. Below are some of the variants which may be relevant to hair loss and treatment.
What Do Your Genetic Results Mean?
Your Result
SRD5A1 (rs248793)
Variant 1 – GG genotype
Variant 2 – CC genotype Variant 3 – GC genotype
What it means At a lower risk of presenting with higher DHT/T ratio At a slightly higher risk of presenting with an increased DHT/T ratio At a higher risk of presenting with a significant increase in DHT/T ratio The Implication You might not be a good candidate for 5α-reductase inhibitors May be a good candidate for normal/increased concentrations of 5α-reductase inhibitors May be a good candidate for increased concentrations of 5α-reductase inhibitors Your Result SRD5A1 (rs3822430)
Variant 1 – AA genotype
Variant 2 – AG genotype Variant 3 – GG genotype
What it means May be a normal/poor responder to 5α-reductase inhibitors May be a good responder to 5α-reductase inhibitors May be a good responder to 5α-reductase inhibitors The Implication May be a good candidate for typical/higher dosages of 5α-reductase inhibitors May be a good candidate for lower dosages of 5α-reductase inhibitors May be a good candidate for lower dosages of 5α-reductase inhibitors Your Result SRD5A1 (rs8192186)
Variant 1 – AA genotype
Variant 2 – AG genotype Variant 3 – GG genotype
What it means May be a good responder to 5α-reductase inhibitors May be a good responder to 5α-reductase inhibitors May be a normal/poor responder to 5α-reductase inhibitors The Implication May be a good candidate for lower dosages of 5α-reductase inhibitors May be a good candidate for lower dosages of 5α-reductase inhibitors May be a good candidate for typical/higher dosages of 5α-reductase inhibitors Your Result SRD5A2 (rs523349)
Variant 1 – GG genotype
Variant 2 – GC genotype Variant 3 – CC genotype
What it means Associated with increased 5α-reductase activity Significantly lower 5α-reductase activity Significantly lower 5α-reductase activity The Implication May be a good candidate for 5α-reductase inhibitors May not be a good candidate for 5α-reductase inhibitors May not be a good candidate for 5α-reductase inhibitors What Relevance Dose SR5DA Have for Hair Loss Treatment?
We have also created a rubric that helps to determine the relevance of a specific gene to hair loss based on the quality of the evidence in the above studies.
On a scale of 1-5, how important are these genetic results? (1 is the lowest, 5 is the highest)
This score is a rating based on evidence quality.
- Does this gene have any potential relevance for hair loss? (1 point)
Yes. Increased DHT is associated with hair follicle miniaturization and the onset of androgenic alopecia, and these genes encode enzymes that synthesize DHT (score=1)
- Does the totality of evidence implicate SRD5A1 or SRD5A2 as causal agents for hair loss? (1 point)
Yes, as mentioned above, an increase in DHT is associated with androgenic alopecia, and these genes create an enzyme that synthesizes DHT (score = 1)
- Does the totality of evidence implicate SRD5A1/SRD5A2 as predictive factors for hair loss treatment responsiveness? (2 points)
Yes, there is evidence to suggest that SRD5A1 may be a predictive factor for hair loss treatment responsiveness, however, some of the patients with the SNP’s for treatment responsiveness still performed poorly, indicating that there are a number of factors involved in responsiveness to treatments. (score = 1)
- Is this quality of evidence on (3) strong enough to influence treatment recommendations? (1 point)
No. (score = 0)
Total Score = 3
Final Thoughts
While some small studies suggest that genetic variation in SRD5A1 and SRD5A2 may influence your response to treatment with a 5α-reductase inhibitor, the evidence is not yet strong enough to make definitive treatment recommendations based solely on genotype. Larger and more robust studies are needed to confirm the true predictive value of genetic testing for SRD5A1 and SRD5A2 for personalizing hair loss treatments.
References[+]
References ↑1 Scaglione, A., Montemiglio, L.C., Parisi, G., Asteriti, I.A., Bruni, R., Cerutti, G., Testi, C., Savino, C., Mancia, F., Lavia, P. and Vallone, B. (2017). Subcellular localization of the five members of the human steroid 5α-reductase family. Biochimie open, 4, 99-106. Available at: https://doi.org/10.1016/j.biopen.2017.03.003 ↑2 Bayne, Flanagan, Einstein, Ayala, Chang, Azzolina, Whiting, Mumford and Thiboutot. (1999). Immunohistochemical localization of types 1 and 2 5α‐reductase in human scalp. British Journal of Dermatology, 141(3), 481-491. Available at: https://doi.org/10.1046/j.1365-2133.1999.03042.x ↑3 Marchetti, P.M. and Barth, J.H. (2013). Clinical biochemistry of dihydrotestosterone. Annals of Clinical Biochemistry, 50(2), 95-107. Available at: https://doi.org/10.1258/acb.2012.012159 ↑4 Gupta, A.K., Bamimore, M.A., Wang, T. and Talukder, M. (2024). The impact of monotherapies for male androgenetic alopecia: A network meta‐analysis study. Journal of Cosmetic Dermatology. Available at: https://doi.org/10.1111/jocd.16362 ↑5, ↑6 Rhie, A., Son, H.Y., Kwak, S.J., Lee, S., Kim, D.Y., Lew, B.L., Sim, W.Y., Seo, J.S., Kwon, O., Kim, J.I. and Jo, S.J. (2019). Genetic variations associated with response to dutasteride in the treatment of male subjects with androgenetic alopecia. Plos one, 14(9), e0222533. Available at: https://doi.org/10.1371/journal.pone.0222533 ↑7, ↑8 Ellis, J.A., Panagiotopoulos, S., Akdeniz, A., Jerums, G. and Harrap, S.B. (2005). Androgenic correlates of genetic variation in the gene encoding 5α-reductase type 1. Journal of Human Genetics, 50(10), 534-537. Available at: https://doi.org/10.1007/s10038-005-0289-x ↑9 Shiota, M., Fujimoto, N., Yokomizo, A., Takeuchi, A., Itsumi, M., Inokuchi, J., Tatsugami, K., Uchiumi, T. and Naito, S. (2015). SRD5A gene polymorphism in Japanese men predicts prognosis of metastatic prostate cancer with androgen deprivation therapy. European Journal of Cancer, 51(14), 1962-1969. Available at: https://doi.org/10.1016/j.ejca.2015.06.122 ↑10 Makridakis, N.M., di Salle, E. and Reichardt, J.K. (2000). Biochemical and pharmacogenetic dissection of human steroid 5α-reductase type II. Pharmacogenetics and Genomics, 10(5), 407-413. Available at: https://doi.org/10.1097/00008571-200007000-00004 Want help with your hair regrowth journey?
Get personalized support, product recommendations, video calls, and more from our researchers, trichologists, and PhD's dedicated to getting you the best possible outcomes.
Learn More
Ben Fletcher, PhD
Benjamin Fletcher is a researcher & writer who holds a BSc in Biological Sciences and an MSc in Genes, Drugs & Stem Cells. Benjamin is currently pursuing a Ph.D. in Molecular Biology & Genetics, conducting research to better understand the regulatory mechanisms that drive muscle atrophy in disease, with a particular focus on the influence of microRNAs.
"... Can’t thank @Rob (PHH) and @sanderson17 enough for allowing me to understand a bit what was going on with me and why all these [things were] happening ... "
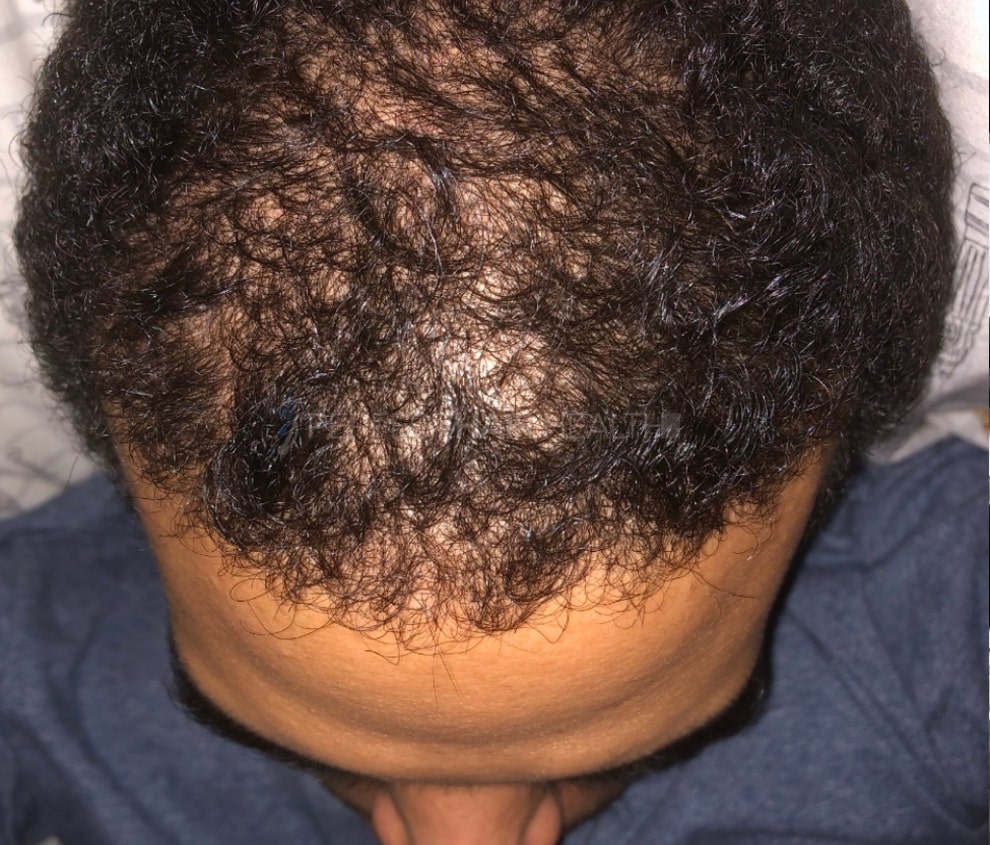
 — RDB, 35, New York, U.S.A.
— RDB, 35, New York, U.S.A."... There is a lot improvement that I am seeing and my scalp feel alive nowadays... Thanks everyone. "
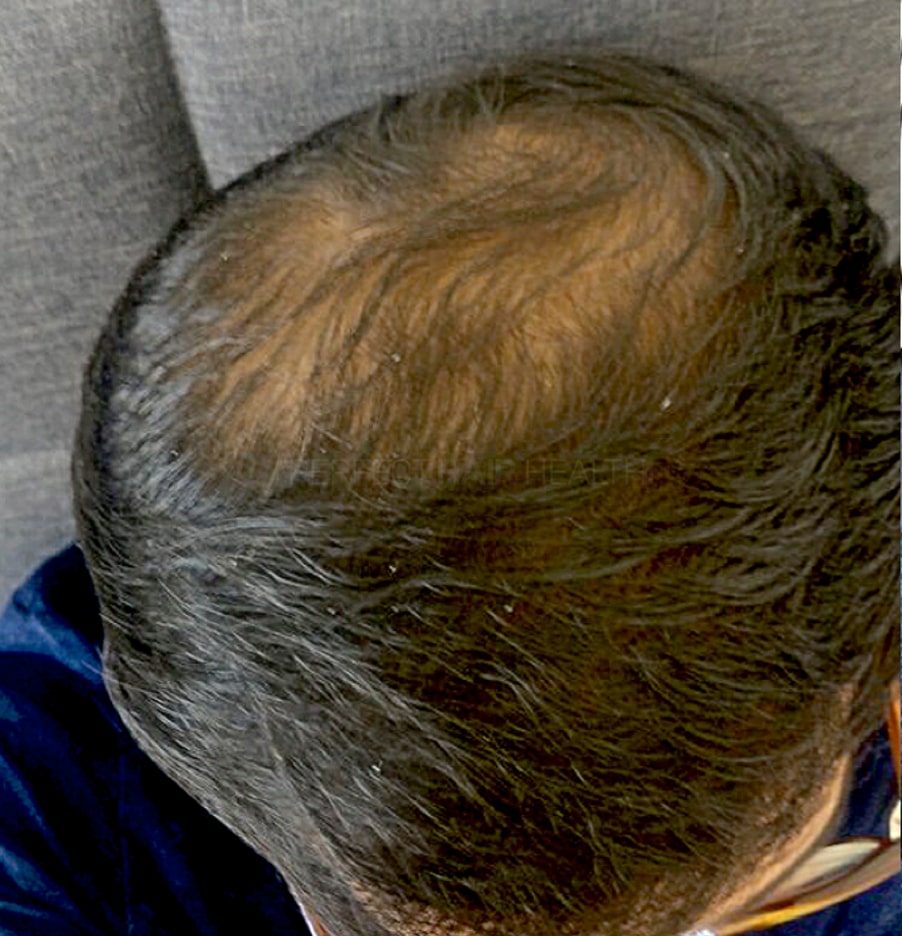
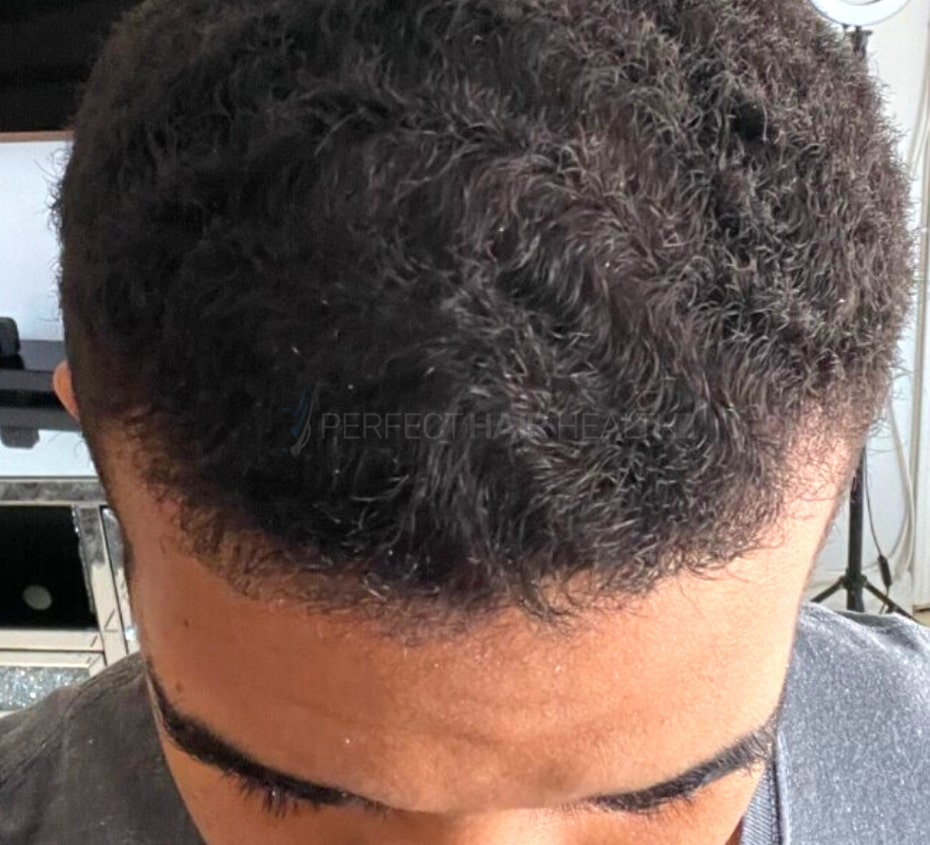
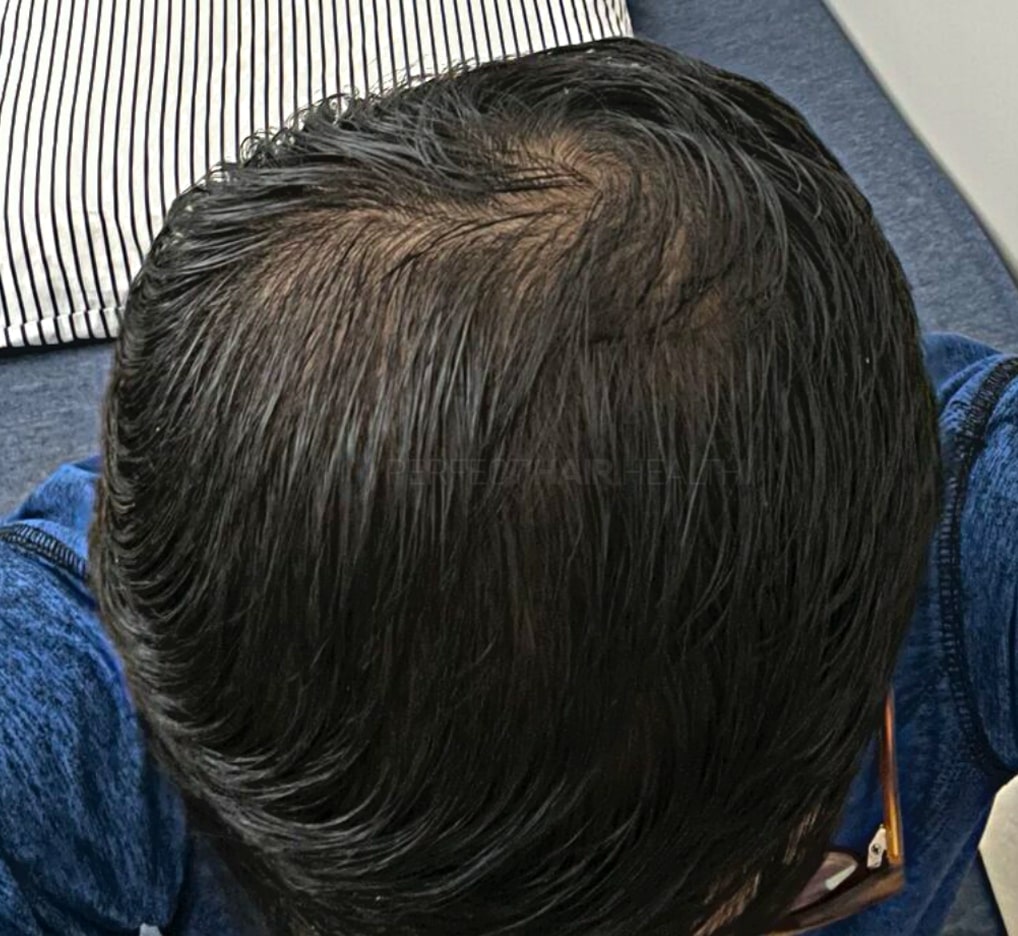 — Aayush, 20’s, Boston, MA
— Aayush, 20’s, Boston, MA"... I can say that my hair volume/thickness is about 30% more than it was when I first started."
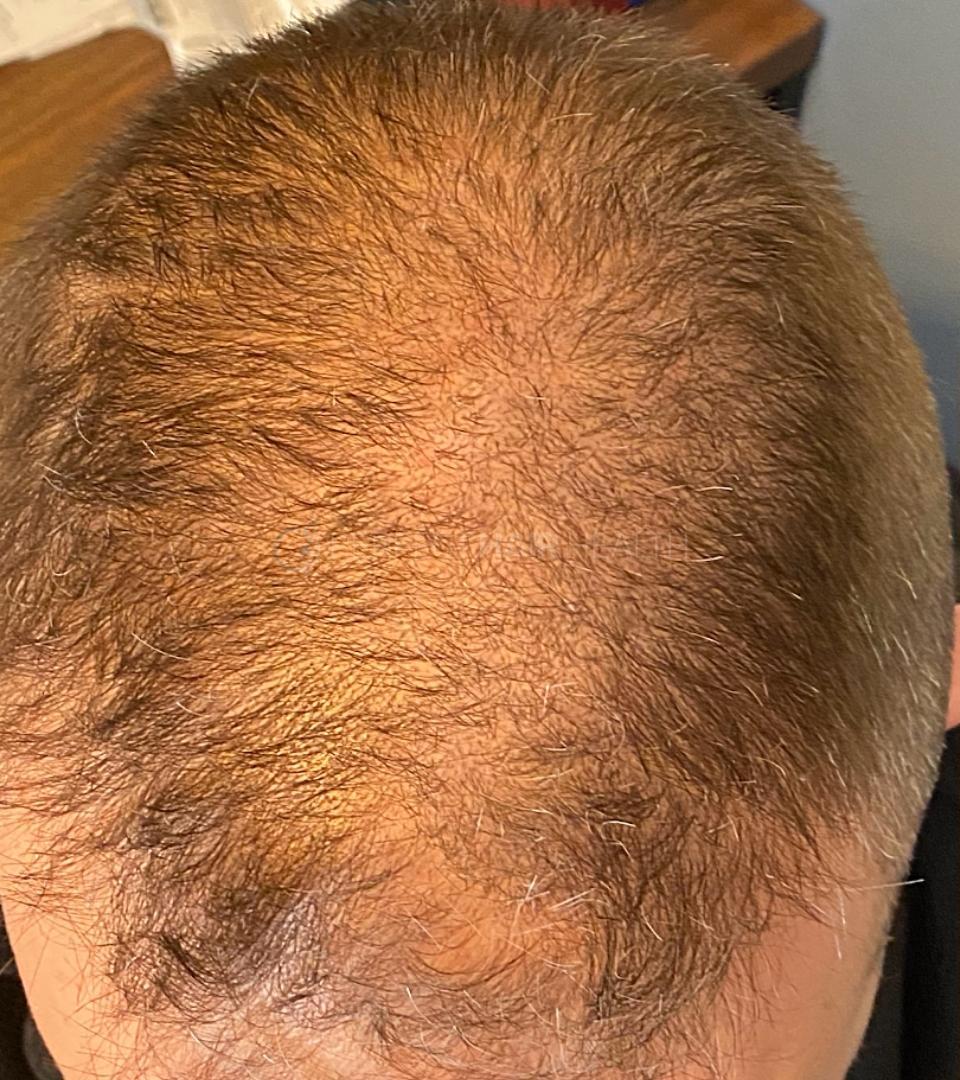
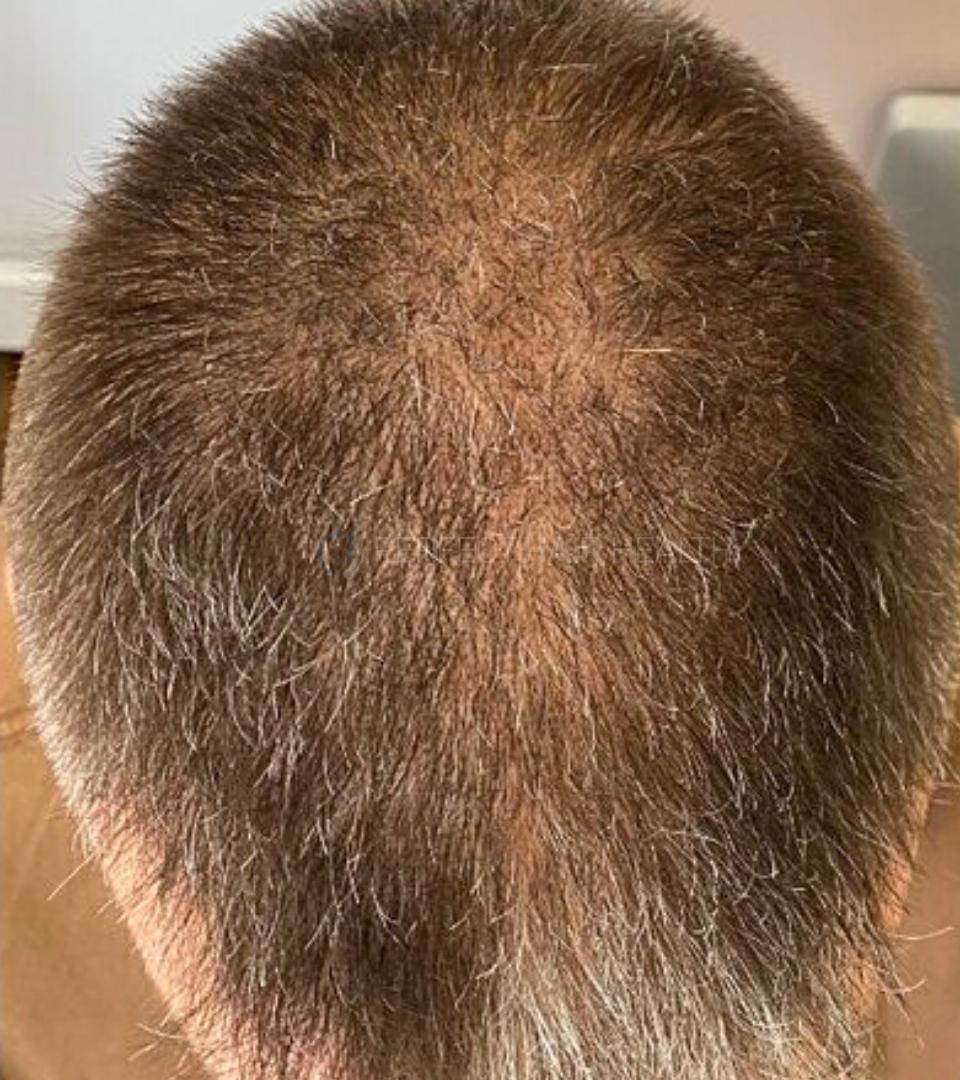 — Douglas, 50’s, Montréal, Canada
— Douglas, 50’s, Montréal, CanadaWant help with your hair regrowth journey?
Get personalized support, product recommendations, video calls, and more from our researchers, trichologists, and PhD's dedicated to getting you the best possible outcomes.
Join Now - Mission Statement